Depending upon the info desired, there are various techniques to examine the products of a PCR response. Agarose gel electrophoresis is a typical method to spot the existence or lack of the target series and the length of the fragment.
Anomaly detection techniques such as denaturing gradient gel electrophoresis (DGGE) and temporal temperature level gel electrophoresis (TTGE) use an acrylamide gel to help with determining mutations in the PCR item.
Semiquantitative analysis strategies allow agarose gel electrophoresis to be used to approximate the starting quantity of material. Sequencing and next-generation sequencing strategy are used when the series of the amplicon must be determined.
Principles
PCR magnifies a specific region of a DNA hair (the DNA target). A lot of PCR approaches amplify DNA fragments of in between 0.1 and 10 kilo base sets (kbp), although some techniques enable amplification of fragments up to 40 kbp in size. The quantity of amplified product is determined by the available substrates in the response, which become limiting as the reaction advances.
A basic PCR set-up requires a number of parts and reagents, consisting of:
- a DNA design template that contains the DNA target area to magnify
- a DNA polymerase, an enzyme that polymerizes brand-new DNA strands; heat-resistant Taq polymerase is particularly typical, as it is more likely to remain intact during the high-temperature DNA denaturation process
- two DNA guides that are complementary to the 3′ (3 prime) ends of each of the sense and anti-sense strands of the DNA target (DNA polymerase can just bind to and lengthen from a double-stranded region of DNA; without primers there is no double-stranded initiation site at which the polymerase can bind); particular guides that are complementary to the DNA target area are picked ahead of time, and are typically customized in a laboratory or purchased from business biochemical providers
- deoxynucleoside triphosphates, or dNTPs (sometimes called “deoxynucleotide triphosphates”; nucleotides including triphosphate groups), the building blocks from which the DNA polymerase manufactures a brand-new DNA strand
- a buffer option supplying an ideal chemical environment for optimal activity and stability of the DNA polymerase
- bivalent cations, generally magnesium (Mg) or manganese (Mn) ions; Mg2+ is the most common, however Mn2+ can be used for PCR-mediated DNA mutagenesis, as a greater Mn2+ concentration increases the mistake rate during DNA synthesis
- monovalent cations, normally potassium (K) ions
The reaction is commonly carried out in a volume of 10– 200 μl in small reaction tubes (0.2– 0.5 ml volumes) in a thermal cycler. The thermal cycler warms and cools the response tubes to achieve the temperatures needed at each step of the reaction.
Lots of contemporary thermal cyclers use the Peltier result, which permits both cooling and heating of the block holding the PCR tubes merely by reversing the electric existing. Thin-walled response tubes allow beneficial thermal conductivity to permit fast thermal equilibration.
Most thermal cyclers have warmed covers to avoid condensation at the top of the response tube. Older thermal cyclers doing not have a heated lid need a layer of oil on top of the reaction mix or a ball of wax inside the tube.
Gel Electrophoresis
PCR products are most frequently examined by agarose gel electrophoresis. The results can be envisioned by ethidium bromide or non-toxic dyes such as SYBR ® green. The intensity of the band can be used to estimate the amount of product of offered molecular weight relative to a ladder. Gel electrophoresis likewise reveals the specificity of the response, where the existence of multiple bands shows secondary amplification products.
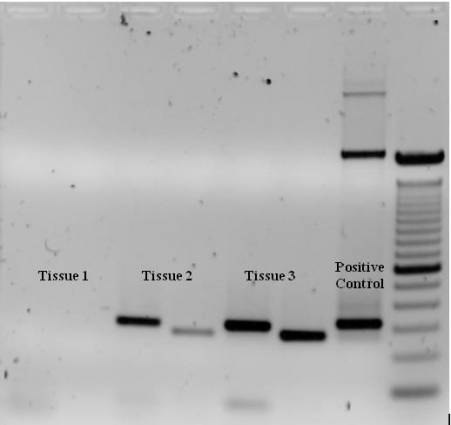
Semiquantitative Analysis
PCR and reverse transcriptase PCR (RT-PCR) are commonly used techniques for discovering species of DNA and RNA, respectively. However the quantitative application of these approaches has been restricted. In theory, PCR magnifies template DNA greatly, with a doubling of design template every cycle, so that relative differences in between samples can be measured as the intensity of bands on the gel.
The restriction is that in later stages of PCR the efficiency reduces and the amplification reaches a plateau. This means that the amount of item in the reaction is no longer proportional to the quantity of starting material and can not be measured. Relative amounts of amplified DNA in different PCR responses can only be compared when the reactions are still in the rapid stage of amplifications. For these reasons, qPCR is the most typically used technique for quantitative analysis.
If appropriate care is required to guarantee that saturation has actually not been reached, end-point PCR can be successfully used as a semiquantitative procedure of relative differences in design template input in between samples.
To guarantee that the response is within the rapid phase of amplification, samples can be magnified with increasing varieties of cycles, work on gel electrophoresis, and quantitated with a gel imager and software. Particular methods for semiquantitative PCR include relative RT-PCR and competitive RT-PCR.
In relative RT-PCR, two primer sets are developed for a multiplexed PCR reaction– one for the gene of interest and another for a housekeeping gene. The primer sets need to be created not to form dimers and to form products of sufficiently different lengths that they can be differentiated through gel electrophoresis.
Pilot experiments must be run to check the product after various numbers of cycles and to figure out the exponential phase of the reaction for both primer sets. Using gel imaging software application to identify the band strength, the relative fold change can then be computed between the samples stabilized to the housekeeping gene.
The difficulty with this technique is that housekeeping genes needed for normalization usually have much higher expression than the gene of interest. One method to this concern is to use lower-expressing housekeeping genes and to use higher concentrations of primers for the gene of interest, and lower concentrations of guides for the housekeeping gene, to favor development of the lower-expressing product.
In competitive RT-PCR, an exogeneous RNA or DNA control is manufactured and increased into the samples. The control should be designed so that it consists of the primer binding sequences as the target however will yield an item of a various length.
The control is then watered down into a basic curve of recognized concentration, and spiked into reproduces of the sample. The exogeneous control and the sample contend for guides and other response components and enhance into two unique bands.
Comparing the strength of the band for the sample to the control of recognized quantity yields a semiquantitative measure of the amount of the records in the sample.
Good luck! Have a nice weekend!
About the Author
Reyus Mammadli is the author of this health blog since 2008. With a background in medical and biotechnical devices, he has over 15 years of experience working with medical literature and expert guidelines from WHO, CDC, Mayo Clinic, and others. His goal is to present clear, accurate health information for everyday readers — not as a substitute for medical advice.